Dockquest
"Claude Code for protein–ligand docking"
Agentic • Automated • Monetizable
doi.bio Agentic DevTools Canada
Steven Ness • sness@sness.net • dockquest.doi.bio
The Problem
- Drug discovery is slow, expensive, failure‑prone (10–15 years; $2.6B; 90% fail).
- Computational tools are being developed rapidly
- Many existing tools and algorithms exist, hard to know what to use and how to use it.
- Much information on how to use these tools and interpret results are locked up in papers.
Team
- Steven R. Ness Ph.D. (papers)
- Serial Founder (Dockvision, folding.ai, docking.ai)
- Background in Software Engineering, Machine Learning, Structural Biology, Rational Drug Design
- CWSF Designer Genes : DockVision : CRANK
- Participated in CASP2, CASP15, CASP16
- Worked as molecular modeller designing drugs
Rational Drug Design
- Pick a validated target (protein) tied to the disease →
- Get its 3D shape (experiment or AI models) →
- Design/score molecules for best “fit” and properties →
- Iterate quickly with wet-lab feedback.
Vision
- Bring new AI tools to drug design
- Help scientists use these new AI tools
- Disrupt all existing drug design software with an agentic model
Moat
- DockQuest Glue + Provenance Store — Hard to rebuild and becomes workflow bedrock.
- doi.bio Literature Graph — Curated paper→residue/ligand links; unique, compounding data asset.
- Learned Reranker + Explainability — Model that re-scores poses using gallery + literature features, producing trusted, regulator-friendly narratives.
Our Insight
- Claude Code pattern is powerful
- Structural Biologists live in CLI and filesystem
- Claude Code lives in the CLI and filesystem
- Run agents where the scientists live
- Give the agents access to the tools, datasets and scientific papers
Solution — What DockQuest Does
- Fold: ESM3/AF‑class structure hypotheses
- Prep: RDKit ligands • protein prep
- Dock: GNINA / AutoDock‑GPU with CNN scoring
- Relax: OpenMM quick minimization
- Explain and Refine: doi.bio literature retrieval
- Reproduce: Inputs, parameters and results saved
CLI‑first; optional web viz (3Dmol.js); MCP/IDE integration; air‑gapped packaging.
Current tools
- Many tools are CLI tools and use the unix filesystem
- Dockquest lives in the unix filesystem and CLI
- Communicate with cloud servers to run jobs in the cloud
Demo
% dockquest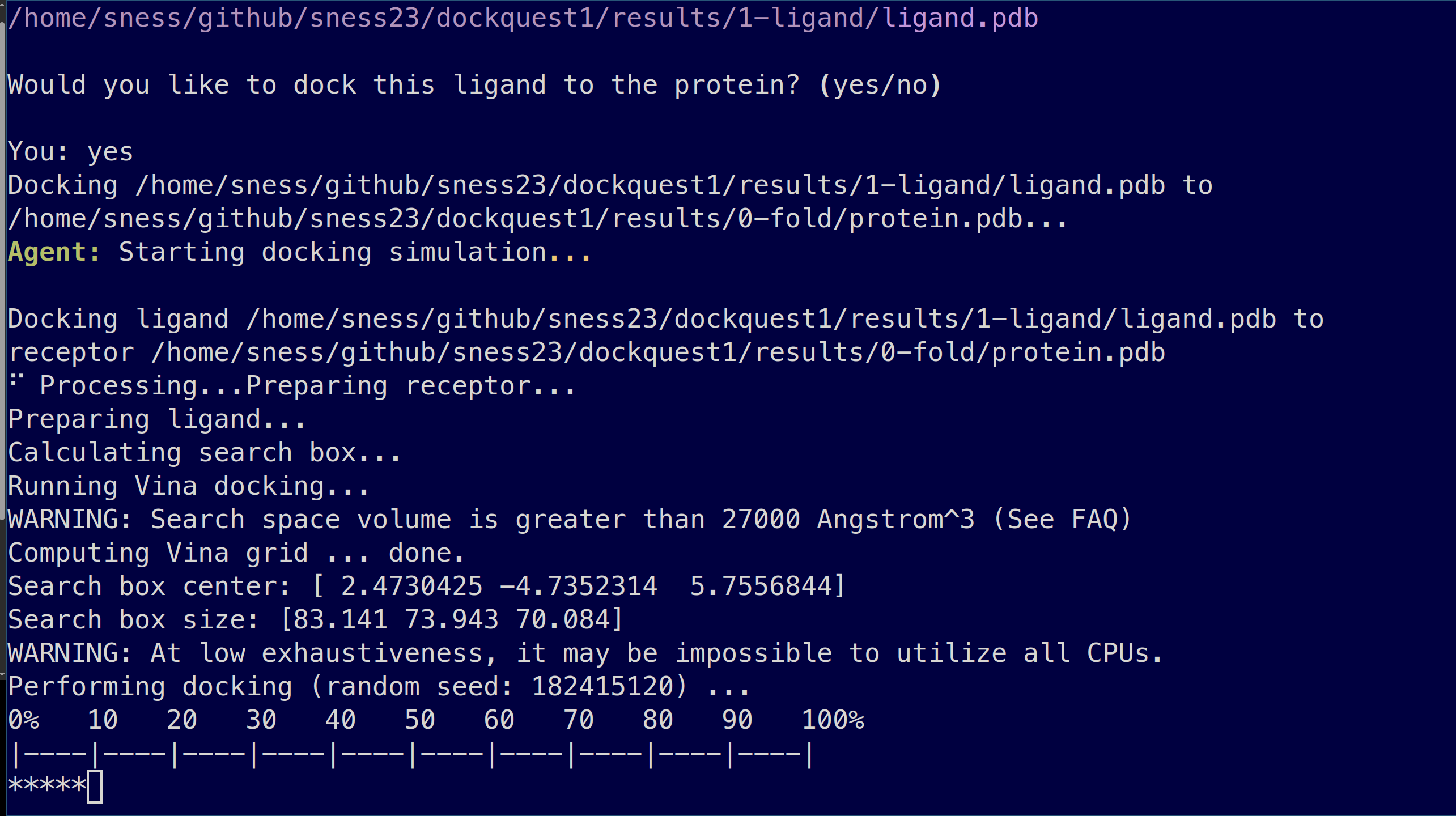
Live 3D — Pose & Pocket
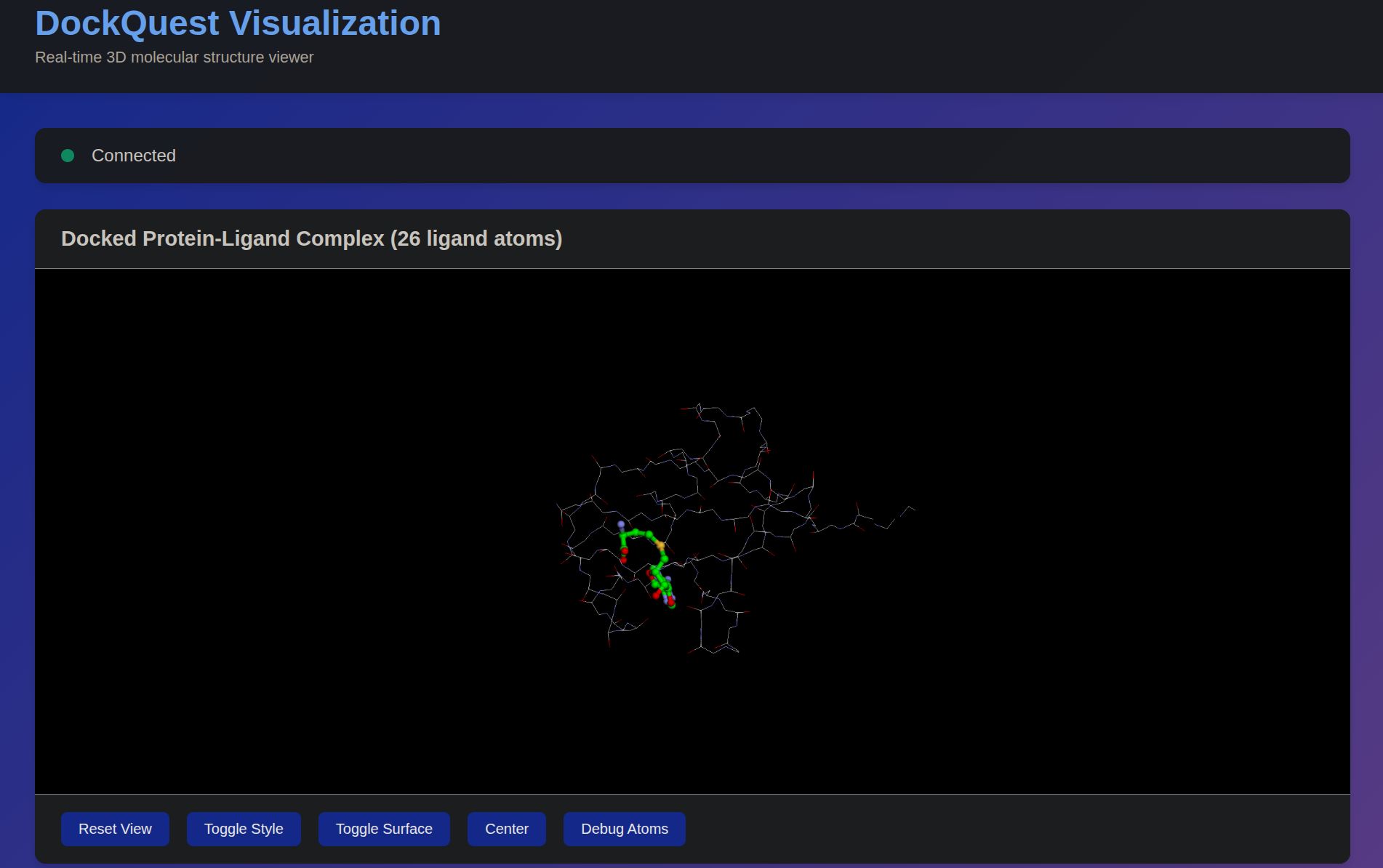
Interactive 3D (3Dmol.js) with surface/pocket highlights, pose gallery, and citations.
Why Now
- ESM3/AF‑class unlocked folding; missing layer is operational glue.
- Vast amount of untapped datasets and scientific papers unlocked
- Apply ideas from Claude Code/Codex/Gemini to Docking
Market
- TAM: $8-12B (SMB biotechs, CROs, academic cores, early pharma).
- SAM: $2.5-5.5B (protein-ligand focus small molecules).
- SOM (5‑yr): ~$50–500M via agentic orchestration + OEM/API.
Business Model & Pricing
- Academic (free + usage)
- Industry Researcher ($99/mo + usage)
- Enterprise (Custom) (~$50K–2M/yr)
- Usage‑metered compute credits • OEM/API licensing • MCP Server
Roadmap (12 Months)
- 0–3 mo: v0.9 agent/adapters/reports • 3 LOIs
- 4–6 mo: MCP Server • Pilots 1–3 live • first conversion
- 7–12 mo: 5 pilots → 2 enterprise • 2 OEM trials • CASP17
Ask (Pre‑Seed)
$500k for 12 months
- 70% GTM‑Science ($350k CAD): 2 Scientist‑BDs
- 20% Compute & Data ($100k CAD): GPUs/CPU, storage, pilots
- 10% Trust & Ops ($50k CAD): legal, SOC2 prep
Contact
Steven Ness • sness@sness.net • dockquest.doi.bio